

















In the Find Best References Using Read Mapping dialog, the 'References' and 'Host references (for filtering)' fields both accept one or more sequence elements, sequence lists, and sequence tracks as inputs, but only the first element in each is used by the tool.
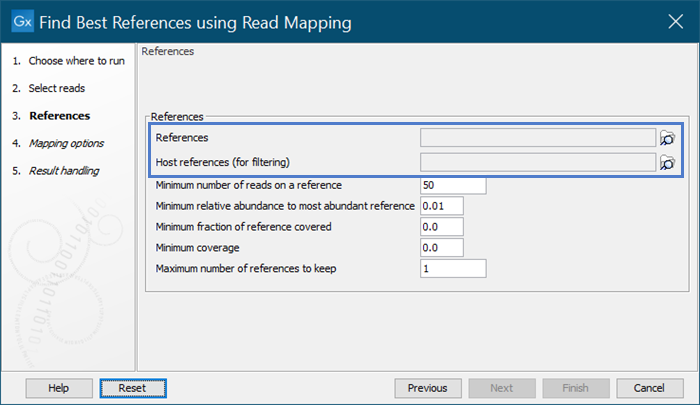
If you provided two or more inputs for ‘References’, results reflect mapping to only the first sequence element, sequence list, or sequence track. Potential better matching references in subsequent input will be missed.
If you provided two or more inputs for the optional ‘Host references (for filtering)’, only reads mapping to the first sequence element, sequence list, or sequence track will be filtered i.e., host read filtering may be incomplete.
Find Best References Using Read Mapping is included in the following Microbial Template Workflow, why results from this workflow will be affected if more than one 'References' or 'Host references (for filtering)' input is provided:
Open results and to go to History View by clicking on the Show History icon under the View area.
Under ‘Parameters’, inputs provided for ‘References’ and ‘Host references (for filtering)’ are given as comma-separated lists. If only one input is listed for either (sequence elements, sequence lists and sequence tracks), your data is not affected by this issue.
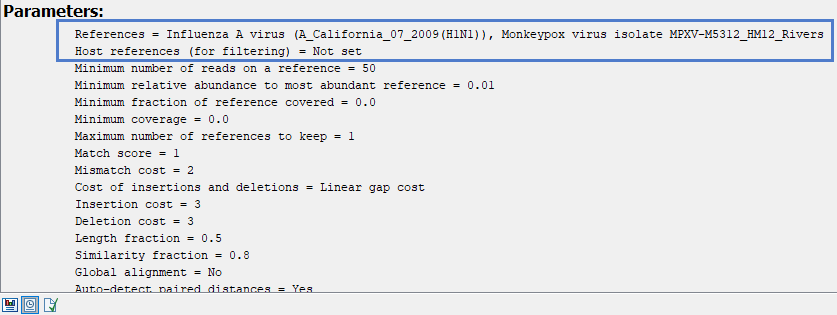
Include all relevant 'References' in one sequence list and provide this as input. Do the same for the optional 'Host references' input.
You can create sequence lists from individual sequences or sequence lists using the Create Sequence List tool:
CLC Microbial Genomics Module 21.1 to 22.1.1.