Issue description
LightSpeed Fastq to Germline Variants includes broken read pairs and ambiguously mapped reads (non-specific matches) when calculating count and coverage, irrespective of tool settings. This can lead to incorrect variant count, coverage and frequency.
The issue does not affect variant calls at single positions or zygosity. However, in very rare cases, joining of adjacent single base pair variants to longer variants can be impacted.
Affected variants:
- When calling variants using the “Ignore non-specific matches” option, all variants overlapping broken or ambiguously mapped reads are affected.
- When calling variants based on both specific and non-specific matches, only variants overlapping broken reads are affected.
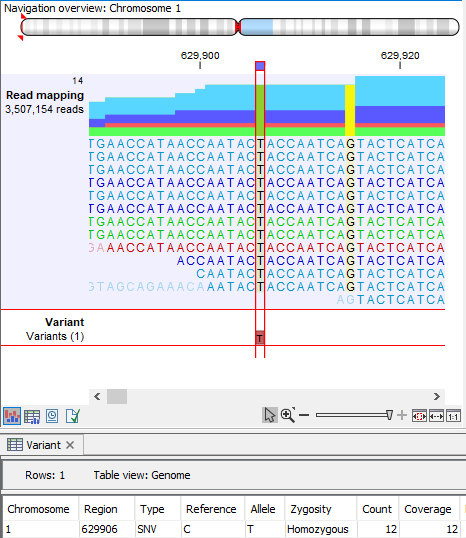
Figure 1. Example of a variant in a region with broken reads (red and green reads). The count and coverage include the broken reads.
Affected software
- CLC LightSpeed Module 23.0
This issue was addressed in CLC LightSpeed Module 23.0.1