

















The Advanced Structural Variant Detection (beta) plugin to the CLC Genomics Workbench is designed to identify structural variants based on combined evidence from unaligned read ends and coverage information of Whole Genome Sequencing (WGS) read mappings. It detects SVs with higher sensitivity and precision than the existing InDels and Structural Variants tool in the standard CLC Genomics Workbench toolbox.
The plugin is simple to use: just install and run on a whole genome sequencing read mapping from within the Workbench.
Known limitations:
Note that this plugin is in beta.
Check out this blog with performance benchmarks of this plugin.
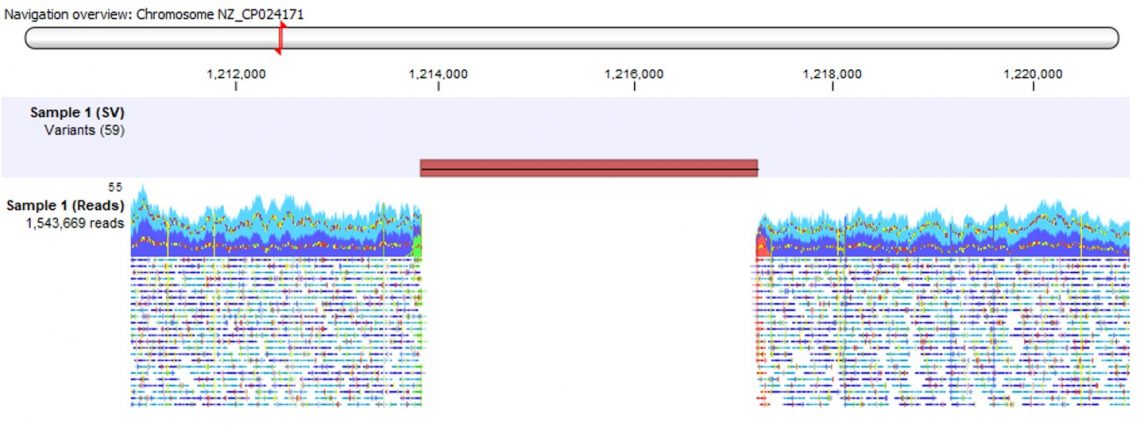
Figure 1. A genome track view of a read mapping and a 3407 bp long deletion identified with the Advanced Structural Variant Detection (beta) plugin.