


















QCI Interpret Translational is a NGS variant assessment software solution that enables rapid, evidence-powered variant annotation, filtering, and triage for human exome, genome, and large cohort sequencing data.
Leveraging the QIAGEN Knowledge Base, QCI Interpret Translational improves research efficiency and accuracy by automating manual curation processes, dynamically and transparently assessing variants according to society guidelines with full user-control, allowing users to focus on what matters most: transforming genomic data into publishable insights.

Eliminate manual curation
Connected to the QIAGEN Knowledge Base, QCI Interpret Translational brings all the information needed for variant assessment together in one location.
Accelerate variant filtering
Leveraging powerful algorithms and trusted content, QCI Interpret Translational applies proprietary filters to rapidly identify potential causal variants within minutes.
Retain full user-control
Unlike other software, QCI Interpret Translational computes assessments with full transparency, giving users control over parameters, policies, and output.

QCI Interpret Translational rapidly identifies the most compelling disease variants in human sequencing data by combining powerful analytical tools and unparalleled content from the QIAGEN Knowledge Base.
The QIAGEN Knowledge Base is the industry’s largest collection of biological and clinical findings, with roughly 2,000,000 unique variants expertly curated from over 300,000 scientific articles, including 140,000 variants connected to the top 200 newborn/carrier screening genes.
Learn more about the QIAGEN Knowledge Base here.
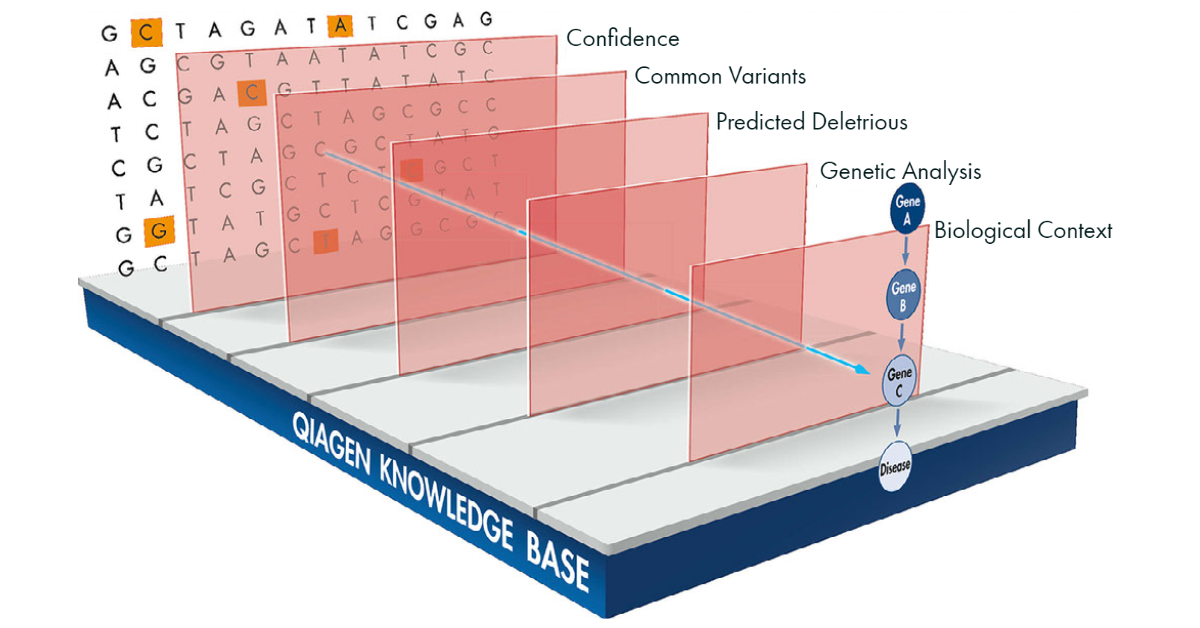
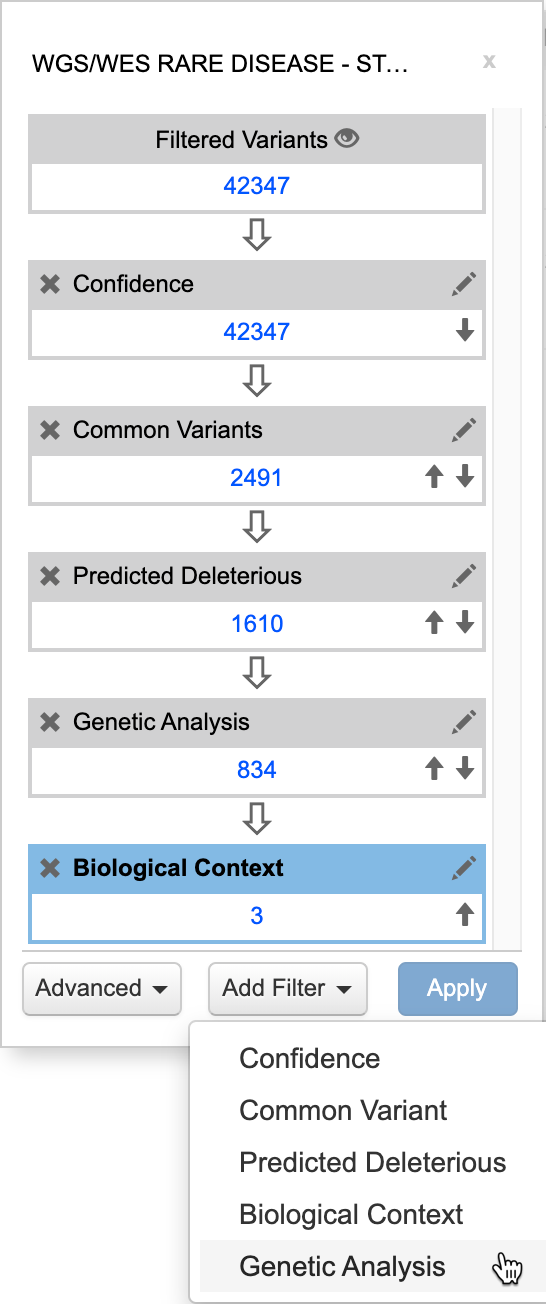
See the Interactive Filter Cascade in action. Watch a video tutorial to learn how to use QCI Interpret Translational.
View the video tutorial here.
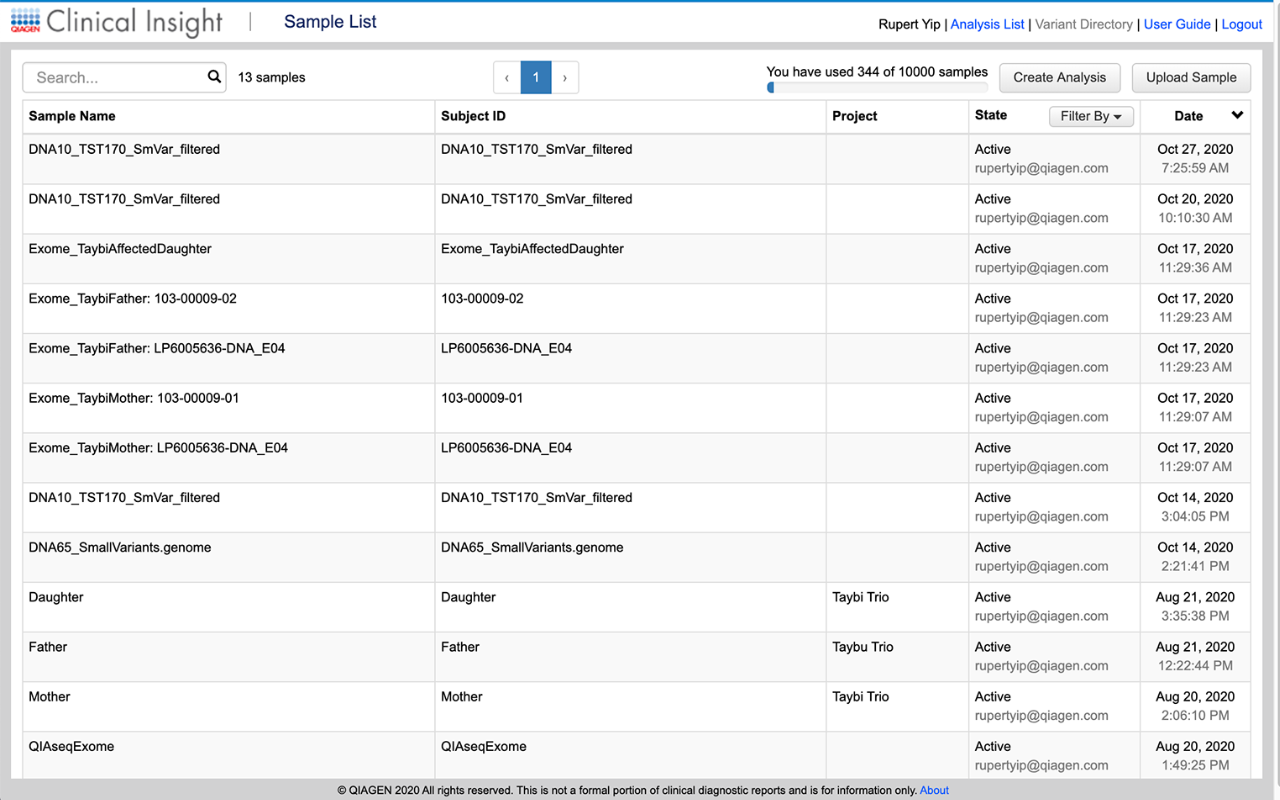
Upload and organize sample data files based on specific projects or by pipeline, and annotate files with detailed descriptions for easy tracking.
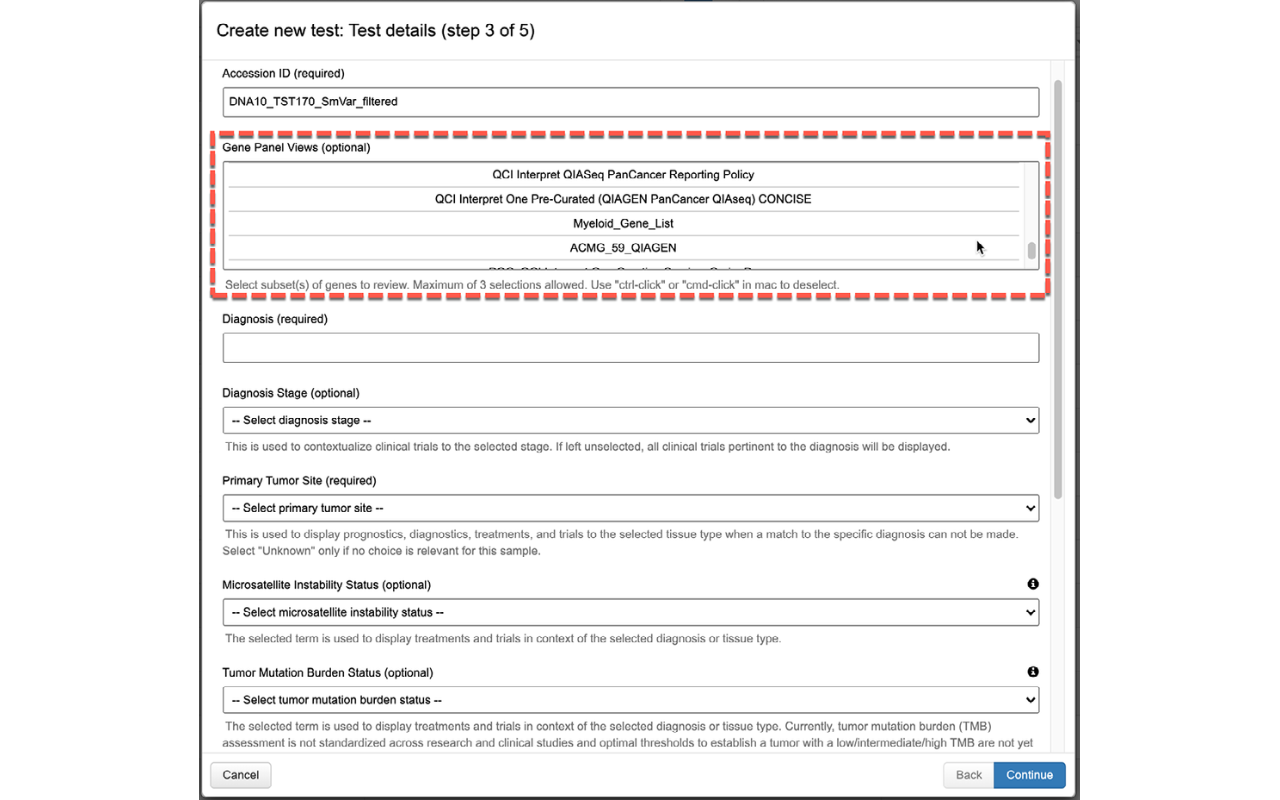
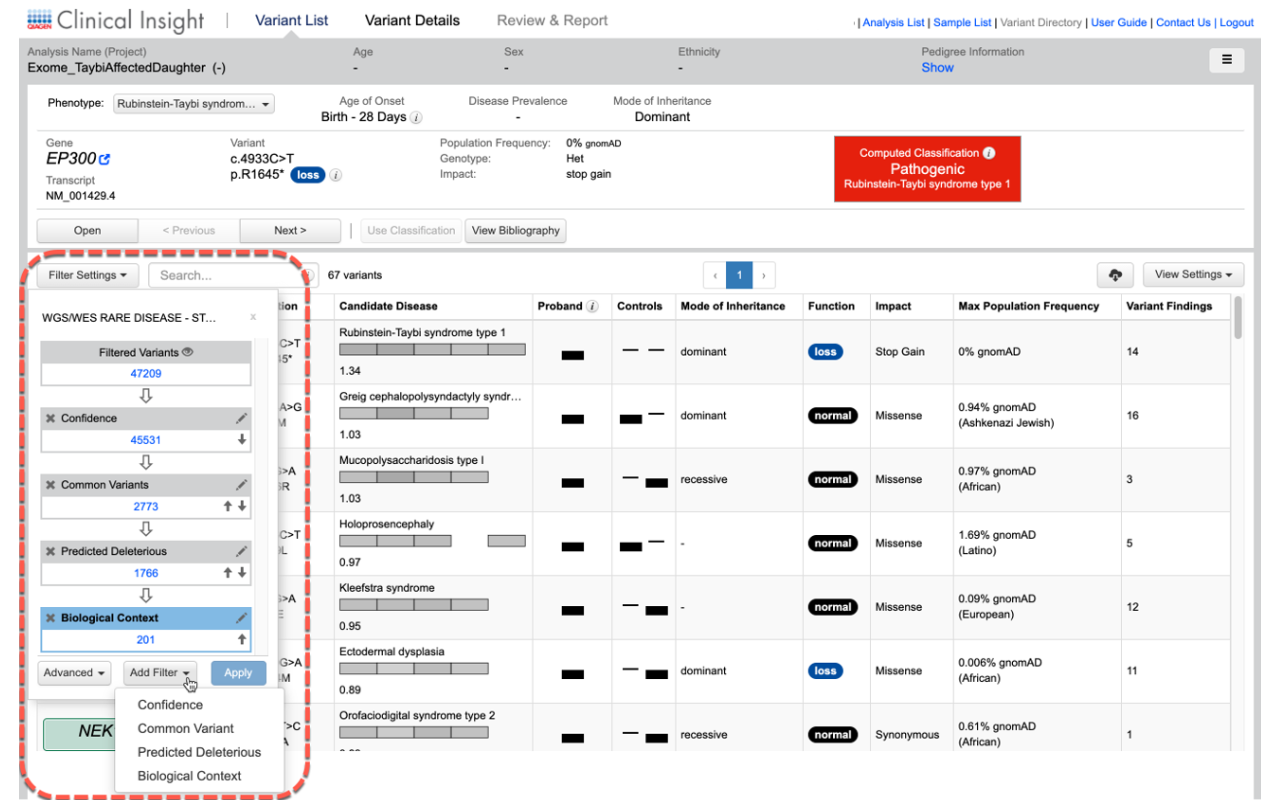
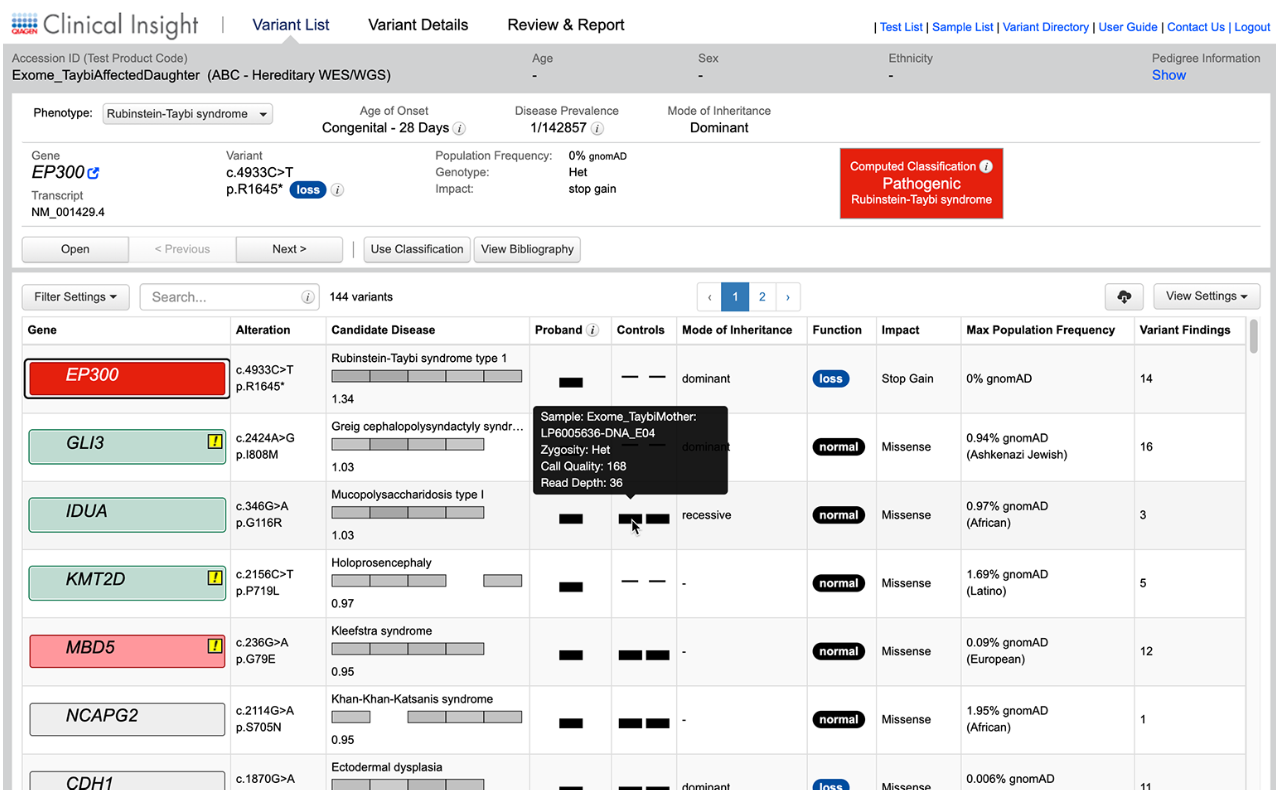



Easily view how and why a variant was classified with the ability to change final assessments depending on your experience and judgment.
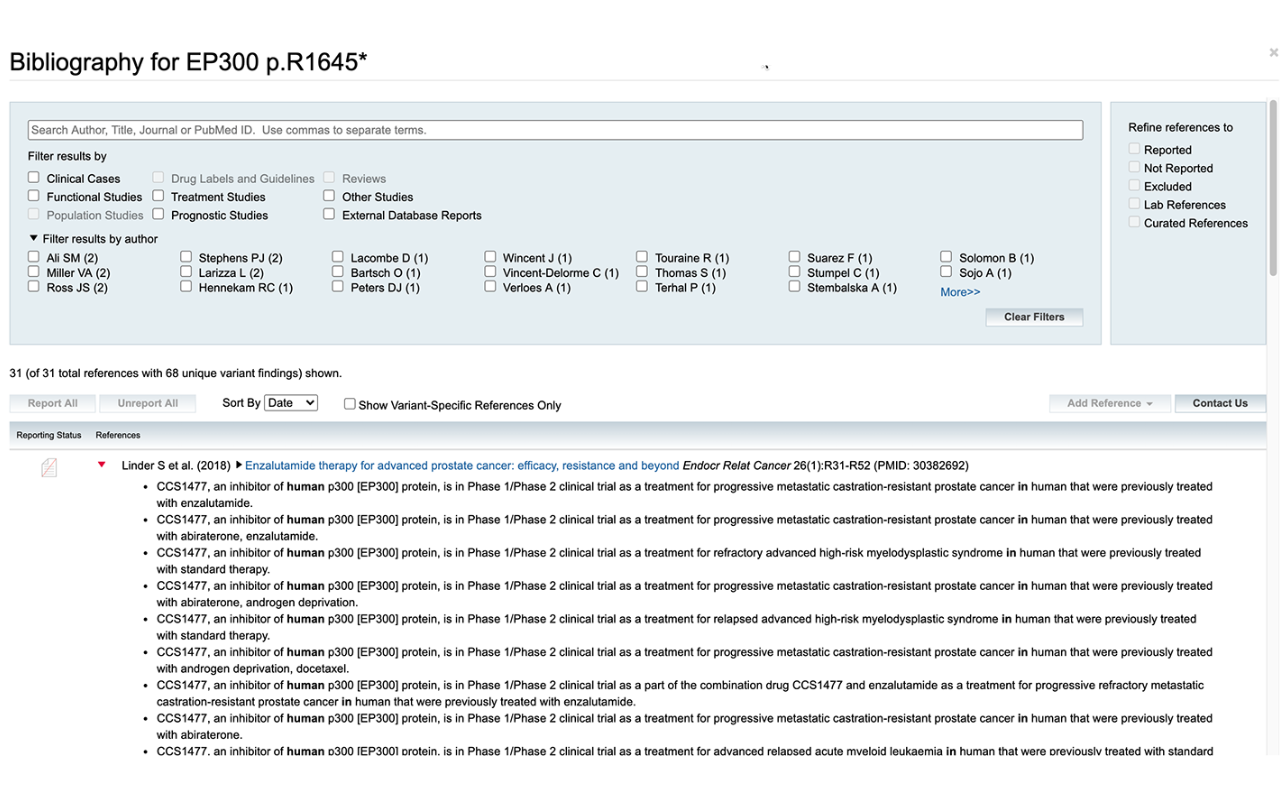

This case study outlines a filtering strategy to identify true, rare, homozygous variants in claudin-low cell lines for functional follow-up in the lab. The video showcases an analysis workflow to search for putatively functional variants in claudin-low breast cancer cell lines by comparing them to luminal breast cancer cell lines using QCI Interpret Translational.

Did you know we offer complimentary trials of our software? No restricted features, no sample data - you get to try all the features of QCI Interpret Translational with your data and see how it works.

Learn more about QIAGEN’s QIAseq Human Exome Kits.


