

















The ClinVar 20220528 reference data elements provided within the QIAGEN Sets "hg19 (Ensembl), version 3" and "hg38 (Ensembl) version 3" (Figure 1) are incomplete, containing only 198 non-reference alleles. More than 1.4 million non-reference alleles should have been present.
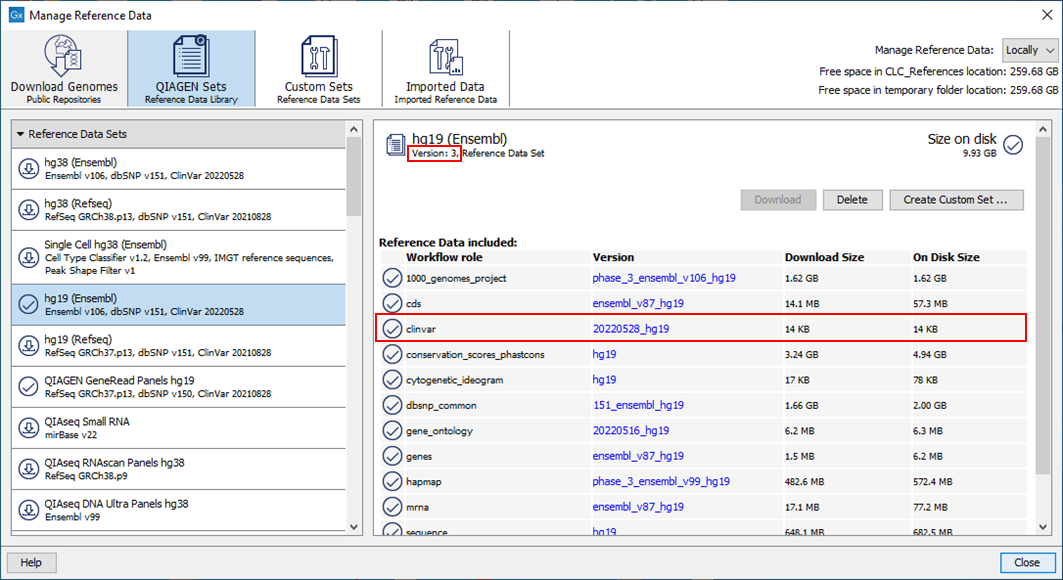
Figure 1. The reference data set hg19 (Ensembl) version 3, where ClinVar is incomplete, as seen in the Reference Data Manager.
If you have downloaded affected reference data sets and have run analyses that include an annotation step making use of Clinvar, relevant variant annotations may be missing in your results.
The affected data elements were released on June 21, 2022, and new, complete elements were released on August 19, 2022. Thus, only hg19 or hg38 Ensembl reference data sets, or Clinvar elements, downloaded between these dates using the Workbench Reference Data Manager are affected.
Some Template Workflows delivered with the Biomedical Genomics Analysis 22.1 plugin, released on June 28, 2022, are pre-configured to use the affected hg19 reference data. Details of the workflows affected are provided below, along with recommended actions to take.
ClinVar databases distributed with other versions of "hg19 (Ensembl)" and "hg38 (Ensembl)" reference data sets are not affected.
The reference data sets were available for download between June 21, 2022, and August 19, 2022, using Genomics Workbench 22.0 and newer. Earlier versions of Genomics Workbench are not affected.
The following Template Workflows provided by the Biomedical Genomics Analysis 22.1 plugin are configured to use the affected reference data:
If you have specified an affected Clinvar data element manually for an analysis, please download an updated data element and re-run your analyses.
Go to References in the top right corner of the CLC Genomics Workbench. Choose QIAGEN Sets and under Reference Data Elements, select a ClinVar reference data element and choose Download. Note that only complete versions of ClinVar are available for download under Reference Data Elements.
If the affected ClinVar reference data elements have been downloaded, they should be deleted. Go to References in the top right corner of the CLC Genomics Workbench. Choose QIAGEN Sets and under Previous Reference Data Elements Sets, select Clinvar 20220528_hg19 or Clinvar 20220528_hg38_no_alt_analysis_set and choose Delete (Figure 2).
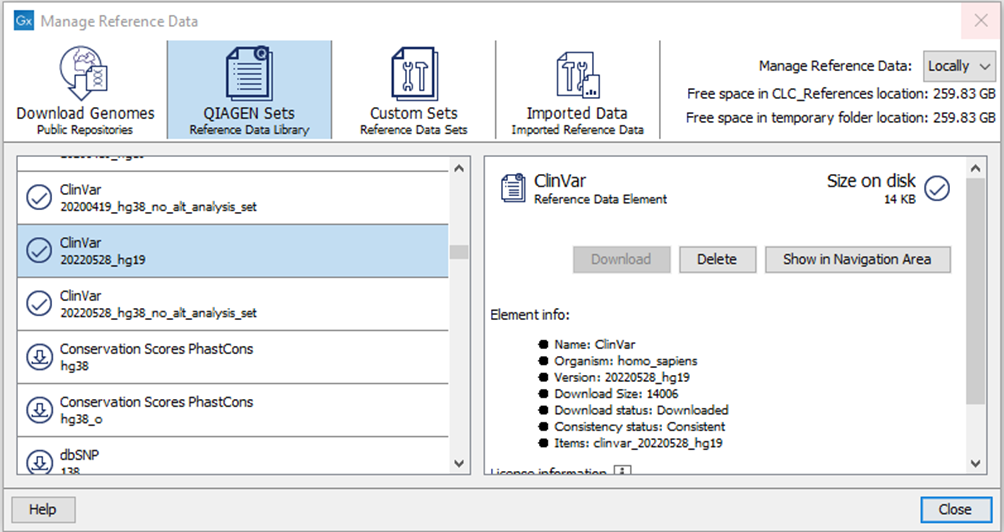
Figure 2. Delete downloaded ClinVar 20220528 reference data elements via the Reference Data Manager.
After a reference data update on August 19, 2022, it is no longer possible to select the affected reference data sets when launching the affected Template Workflows.
However, if using Biomedical Genomics Analysis plugin 22.1, and the affected hg19 Ensembl reference set has been downloaded, and an affected Template Workflow is launched with the option “Use the default reference data” selected, the incomplete clinvar_20220528_hg19 reference data element will be used to annotate variants unless the action described below is taken.
If you have the Biomedical Genomics Analysis 22.1 plugin installed and have downloaded the affected reference data for hg19
Delete the affected ClinVar reference data element following the steps described above. If the incomplete ClinVar element is not deleted, by default, affected Template Workflows will suggest “Use the default reference data” when launched (Figure 3). If checking this option, the workflow will proceed using the incomplete ClinVar database.
If you choose not to delete the incomplete ClinVar reference data element, you can avoid using it by checking “Select a reference set to use” and choosing one of the reference data sets under QIAGEN Active or QIAGEN previous (Figure 3).
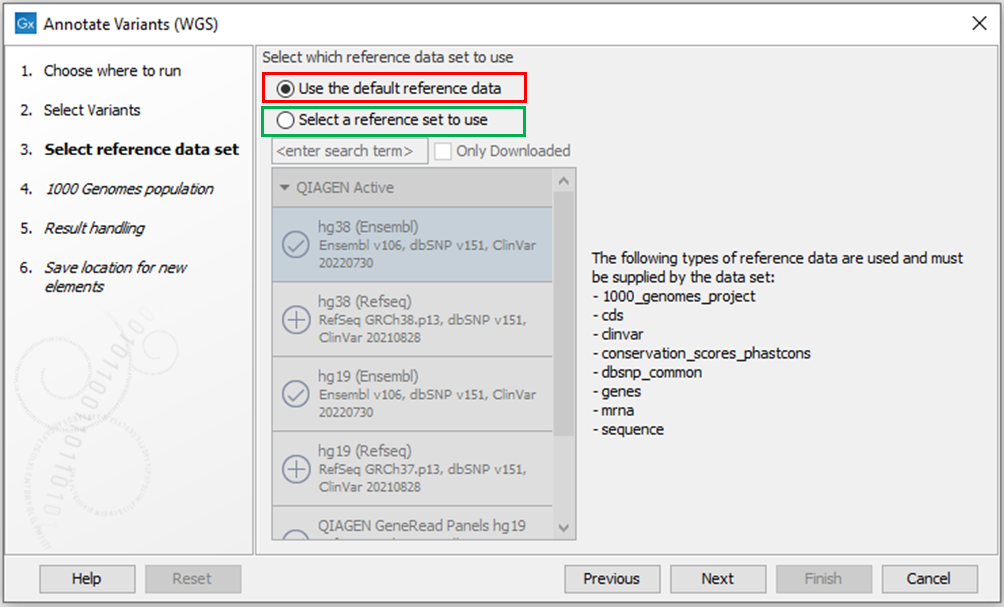
Figure 3. When clinvar_20220528_19 has been downloaded, affected workflows will suggest “Use the default reference data” by default (red box). Instead choose “Select a reference set to use” to specify one of the reference data sets, which all contain complete ClinVar databases (green box).
Other situations
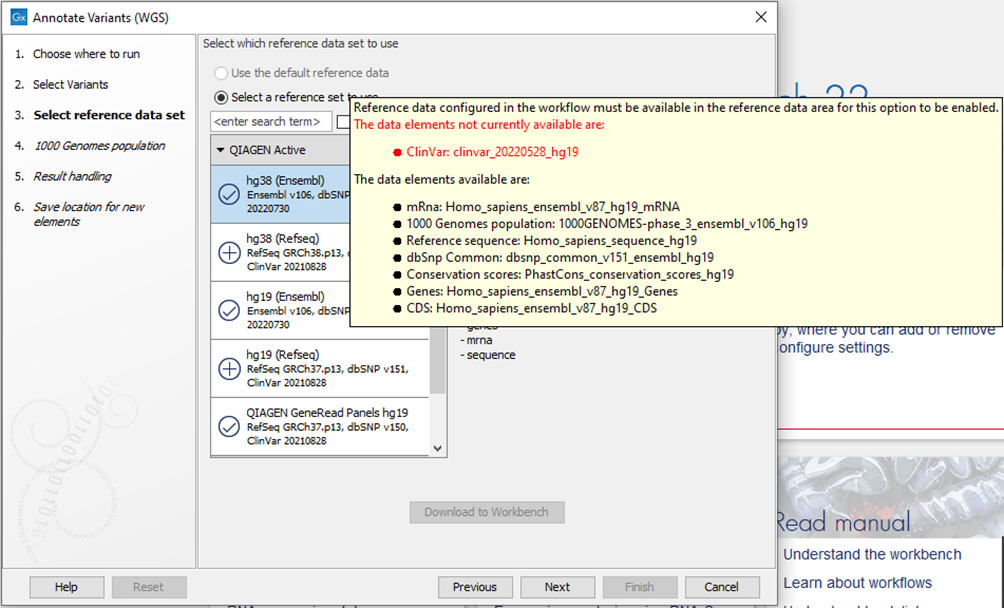
Figure 4. Affected Template Workflows using the ClinVar 20220528 database can no longer be run using default reference data.