

















Differential Abundance Analysis is based on sample metadata. In the tool dialog, you select the metadata factor that you wish to analyze. Samples are grouped based on their values for this factor, and the algorithm evaluates how the abundances of features (e.g., species or genera) differ between these groups.
When the underlying samples have more than one metadata association, if you select a metadata factor from metadata association two, three, or later, the algorithm wrongfully uses the values from metadata association one. Consequently, samples are grouped on the wrong values, and the analysis is carried out with the wrong sample grouping.
Open the underlying samples and go to Elements Info View by clicking on the Show Elements Info icon under the View area. Under ‘Metadata’, check to see if you have more than one metadata association.
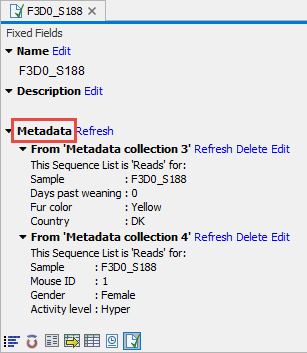
If you see only one metadata association, your differential abundance analysis is not affected by this issue.
If you see two or more metadata associations, you need to further check the factor on which your differential abundance analysis is based.
If your analysis is based on a factor from the first metadata association, your differential abundance analysis is not affected by this issue.
If, however, your differential abundance analysis is based on a factor from metadata association two, three, or later, your differential abundance analysis is most likely affected.
To look up the factor on which your differential abundance analysis is based, open the differential abundance analysis, to go to History View by clicking on the Show History icon under the View area. You find ‘Metadata factor’ under ‘Parameters’.
With the above example, if my differential abundance analysis is based on the factor ‘Fur color’, my results are not affected. If, however, the applied factor is ‘Gender’, my differential abundance analysis will be affected.
If your differential abundance analysis is affected by this issue, we recommend that for the involved samples, you delete all metadata associations except the one relevant for your differential abundance analysis and rerun your full analysis pipeline.
To delete metadata associations, open the samples one by one, and go to Elements Info View by clicking on the Show Elements Info icon under the View area. Click ‘Delete’ next to the metadata associations that you wish to remove and click Save
This issue affects CLC Microbial Genomics Module 22.1.1 and earlier, and 23.0.
This issue was addressed in CLC Microbial Genomics Module 22.1.2 and 23.0.1.